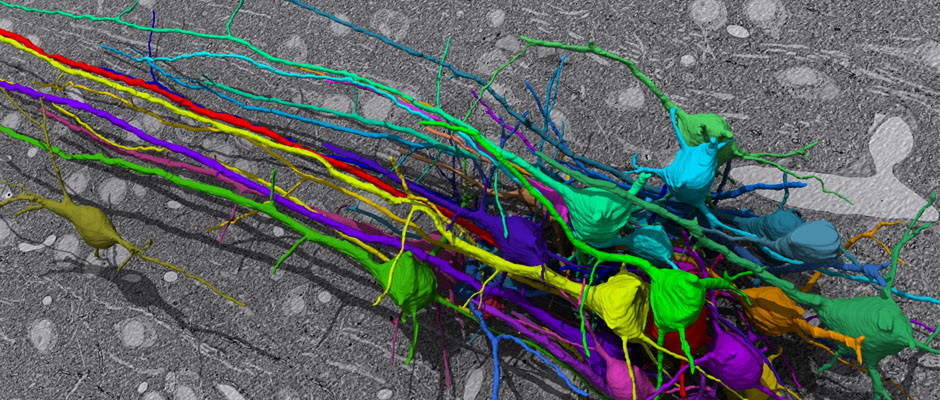
SNEMI3D: 3D Segmentation of neurites in EM images (ISBI 2013)¶
(This website is the replacement of the old one)
Welcome to the server of the first challenge on 3D segmentation of neurites in EM images! The challenge is organized in the context of the IEEE International Symposium on Biomedical Imaging (San Francisco, CA, April 7-11th 2013).
Background and set-up¶
In this challenge, a full stack of electron microscopy (EM) slices will be used to train machine-learning algorithms for the purpose of automatic segmentation of neurites in 3D. This imaging technique visualizes the resulting volumes in a highly anisotropic way, i.e., the x- and y-directions have a high resolution, whereas the z-direction has a low resolution, primarily dependent on the precision of serial cutting. EM produces the images as a projection of the whole section, so some of the neural membranes that are not orthogonal to a cutting plane can appear very blurred. None of these problems led to major difficulties in the manual labeling of each neurite in the image stack by an expert human neuro-anatomist.
In order to gauge the current state-of-the-art in automated neurite segmentation on EM and compare between different methods, we are organizing a 3D Segmentation of neurites in EM images (SNEMI3D) challenge in conjunction with the ISBI 2013 conference. For this purpose, we are making available a large training dataset of mouse cortex in which the neurites have been manually delineated. In addition, we also provide a test dataset where the 3D labels are not available. The aim of the challenge is to compare and rank the different competing methods based on their** object classification accuracy** in three dimensions.
The image data used in the challenge was produced by Lichtman Lab at Harvard University (Daniel R. Berger, Richard Schalek, Narayanan "Bobby" Kasthuri, Juan-Carlos Tapia, Kenneth Hayworth, Jeff W. Lichtman) and manually annotated by Daniel R. Berger. Their corresponding biological findings were published in Cell (2015).
The training/test data is hosted on Zenodo.
Results format¶
The results are expected to be uploaded in the challenge server as a a zip file containing a single h5 file named "test-input.h5" containing a volume dataset named "main". Pixels with the same value (ID) are assume to belong to the same object in 3D.
The results are evaluated using the adapted_rand metric.
Organizers¶
- Ignacio Arganda-Carreras, Massachusetts Institute of Technology.
- H. Sebastian Seung, Massachusetts Institute of Technology.
- Ashwin Vishwanathan, Massachusetts Institute of Technology.
- Daniel R. Berger, Massachusetts Institute of Technology.
The image data used in the challenge was produced in Lichtman Lab at Harvard University (Daniel R. Berger, Richard Schalek, Narayanan "Bobby" Kasthuri, Juan-Carlos Tapia, Kenneth Hayworth, Jeff W. Lichtman) and manually annotated by Daniel R. Berger. Their corresponding biological findings were published in Cell (2015).
The organizers thank Alessandro Guisti, Dan Ciresan, Luca M. Gambardella and Juergen Schmidhuber from the Swiss AI Lab IDSIA (Istituto Dalle Molle di Studi sull'Intelligenza Artificiale) for providing the 2D membrane probabilities of the challenge data sets.
The organizers thank Jason Adhinarta and Donglai Wei from Boston College for porting the challenge to grand-challenge for continuous benchmarking.